AI & Computational Chemistry

Where Expert Insight Meets Intelligent Innovation
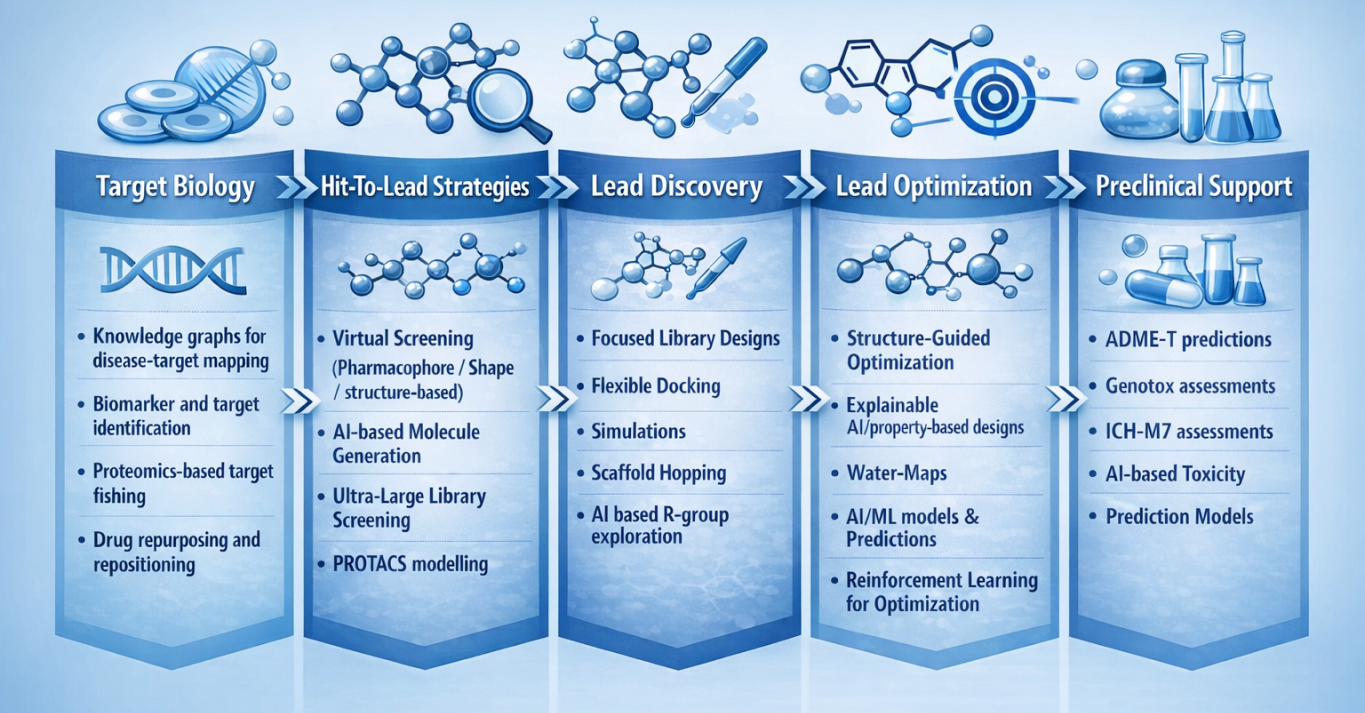
AI & Computational Chemistry at PI Health Sciences unites deep human scientific expertise with advanced artificial intelligence to transform how drugs are designed and optimized. Our AI & Computational Chemistry platform accelerates molecular discovery from target identification and virtual screening through hit-to-lead and lead optimization, enabling pharmaceutical and biotech partners to discover better molecules faster. By embedding safety-aware design principles early, we ensure computational insights directly support confident discovery decisions.
Our multidisciplinary team of computational chemists, medicinal chemists, and AI scientists brings decades of experience in designing clinical candidates across multiple therapeutic areas. Through the integration of deep learning models, molecular simulations, knowledge graphs, and explainable AI frameworks, AI & Computational Chemistry at PI Health Sciences translates complex biological and chemical data into precise, actionable insights. These insights guide rational molecular design, prioritization, and optimization at every stage of modern discovery workflows.
Built as an integrated, scalable platform, AI & Computational Chemistry at PI Health Sciences supports hit-to-lead optimization, structure- and ligand-based design, ultra-large library screening, and early ADMET and genotoxic risk assessment including ICH M7–aligned evaluations. By combining computational rigor with experimental relevance, AI & Computational Chemistry enables faster convergence on viable chemical matter, improved predictability, and higher-quality candidates ready for downstream progression.
Core Services
Adaptive Machine Learning for Data-Efficient Discovery Decisions
Active Learning at PI Health Sciences delivers active learning–driven computational chemistry solutions to accelerate decision-making across drug discovery and early chemical development. By integrating machine learning with expert-in-the-loop decision frameworks, active learning enables predictive models to iteratively identify the most informative compounds or experiments for validation. This approach allows discovery teams to navigate vast chemical space efficiently, reduce experimental burden, and improve decision confidence before committing to large-scale synthesis, screening, or development activities.
How Our Approach Is Different
Active learning at PI Health Sciences is engineered as a scientifically governed, closed-loop decision system, not a black-box prediction engine. Chemical and experimental data are carefully curated to define the learning space, and model outputs are reviewed by experienced computational and medicinal chemists to ensure feasibility, synthetic accessibility, and alignment with program objectives. Experimental results are continuously reintegrated into the workflow, allowing models to evolve based on real data and ensuring traceability as programs progress from discovery into development.
Core Design Principles
- Iterative, closed-loop learning driven by real experimental data
- Explicit identification and use of model uncertainty to guide decisions
- Focus on prioritizing informative experiments rather than maximizing predictions
- Integration of expert scientific review at every decision stage
- Data-efficient learning designed for sparse and evolving datasets
- Alignment with discovery and early development workflows
Talk to our experts
Rational Engineering of Therapeutic and Conjugated Peptides
AI-driven peptide design at PI Health Sciences enables the rational engineering of next-generation therapeutic peptides, peptide–drug conjugates, and epitope-based constructs for advanced biologic programs. By integrating generative AI, structure prediction, binding optimization, and developability scoring, we design peptides that are structurally informed, biologically relevant, and development-ready. Our platform supports peptide discovery across multiple modalities, including protein–protein interaction modulation, targeted delivery, immunotherapy, and peptide-based hybrid formats, translating computational designs into experimentally actionable candidates.
How Our Approach Is Different
Our peptide design workflows are structure-first and decision-driven, not sequence-only generation exercises. AI models are trained to learn peptide sequence–structure–function relationships and are applied within clearly defined biological objectives. Predicted peptides-protein complexes are evaluated using docking, free-energy scoring, and stability analysis, while additional predictive layers assess solubility, aggregation risk, protease resistance, permeability, immunogenicity, and formulation compatibility. All AI outputs are reviewed by experienced scientists to ensure structural plausibility, synthetic feasibility, and alignment with program goals.
Core Design Principles
- Structure-aware peptide generation informed by 3D binding environments
- Focus on relative prioritization of peptide candidates rather than isolated affinity metrics
- Early integration of stability, solubility, and protease resistance assessment
- Design compatibility with conjugation, stabilization, and hybrid biologic formats
- Explicit evaluation of synthetic feasibility and formulation constraints
- Interpretable outputs aligned with peptide chemistry and biology
Talk to our experts
Explainable AI for Rational and Development-Ready Design
AI-powered lead optimization at PI Health Sciences enables data-driven and explainable decision-making during the most design-intensive phase of drug discovery. By integrating explainable artificial intelligence, multi-dimensional structure–property analysis, and predictive ADMET intelligence, we support chemists in making rational, synthesizable, and biologically relevant compound modifications. This approach accelerates convergence toward development-ready leads while minimizing hidden risk accumulation during optimization.
How Our Approach Is Different
Unlike traditional black-box machine learning approaches, PI Health Sciences applies explainable AI methodologies that reveal how specific atoms, functional groups, and structural features influence potency, selectivity, permeability, solubility, and metabolic liability. All AI-generated insights are interpreted by experienced medicinal chemists to ensure chemical feasibility, strategic relevance, and alignment with program objectives. Predictive intelligence is embedded directly into medicinal chemistry workflows, supporting informed Design–Make–Test–Analyze decisions rather than retrospective analysis.
Core Design Principles
- Explainable, atom-level and fragment-level interpretation of SAR and SPR
- Focus on relative ranking and decision guidance rather than raw prediction
- Early integration of ADMET and safety intelligence
- Balanced multi-parameter optimization across potency and developability
- Chemist-reviewed AI outputs to ensure synthetic realism
- Iterative learning integrated into DMTA cycles
Talk to our experts
Generative Design of Novel and Decision-Ready Chemical Matter
AI-driven molecular generation at PI Health Sciences enables the rational design of novel chemical entities before they exist in any database. By leveraging generative artificial intelligence, we move discovery programs beyond screening known chemical space toward the intentional creation of molecules engineered to meet defined biological, physicochemical, developability, and IP objectives. This capability supports focused, decision-ready molecular designs that integrate seamlessly with chemistry, biology, and downstream experimental execution.
How Our Approach Is Different
Molecular generation at PI Health Sciences is executed as a target-aware, medicinal chemistry–led design process, not a black-box ideation exercise. Design objectives are explicitly defined upfront and translated into multi-objective reward functions that guide generative models toward chemically relevant solutions. All generated outputs are interpreted by experienced medicinal and computational chemists to ensure chemical plausibility, synthetic tractability, and strategic alignment before any synthesis commitment is made.
Core Design Principles
- Explicit definition of biological, physicochemical, and developability objectives
- Multi-objective optimization balancing potency, selectivity, ADMET, and novelty
- Exploration of high-novelty, IP-aware chemical space beyond existing libraries
- Integration of synthetic feasibility and retrosynthesis considerations early
- Human-in-the-loop interpretation to ensure chemical realism and relevance
- Seamless alignment with downstream synthesis and experimental workflows
Talk to our experts
Rapid Hit Discovery Across Billion-Scale Chemical Space
Ultra-Large Library Screening at PI Health Sciences enables rapid, data-driven hit discovery from expansive chemical spaces containing billions of molecules. By combining AI-driven ligand-based and structure-based virtual screening with large-scale computational infrastructure, we help pharmaceutical and biotech programs identify high-quality, synthesis-ready hits in days rather than months. This capability allows discovery teams to access unprecedented chemical diversity early, without compromising precision, novelty, or downstream feasibility.
How Our Approach Is Different
Ultra-large library screening at PI Health Sciences is executed as a scientifically governed, multi-stage decision process, not a single-pass virtual screen. Libraries sourced from commercial vendors, make-on-demand inventories, public databases, and curated internal collections are systematically filtered for chemical validity, liability risk, and novelty. Screening strategies are selected based on data availability, combining ligand-based similarity, pharmacophore alignment, and GPU-accelerated structure-based docking. All shortlisted hits are reviewed by experienced scientists to ensure interpretability, relevance to the biological hypothesis, and readiness for synthesis and follow-on optimization.
Core Design Principles
- Exploration of billion-scale chemical space with staged refinement
- Strategic use of ligand-based and structure-based screening workflows
- Early filtering for chemical quality, liabilities, and redundancy
- Multi-parameter hit prioritization beyond docking scores alone
- Emphasis on synthetic feasibility and make-on-demand readiness
- Expert scientific review to ensure decision relevance and clarity
Talk to our experts
Capabilities
Structure & Ligand-Based Design
We use advanced protein modelling, pharmacophore mapping, and GPU-accelerated molecular docking to identify synthetically feasible scaffolds with high-affinity, selective target interactions.

AI-Augmented Hit-to-Lead Optimization
Our AI-driven design loops integrate generative models, deep learning scoring, and explainable AI to rapidly prioritize synthetically tractable compounds with optimal potency and selectivity.
Active Learning & Ultra-Large Library Screening
Active learning–driven virtual screening enables efficient exploration of billion-scale libraries, refining diverse, high-affinity hits with minimal computational overhead.

Synthons-Driven Design & Enumeration
PI Health Sciences applies reaction-based enumeration and building block intelligence to generate novel, synthetically accessible compounds that seamlessly translate from AI design to laboratory execution.

Mechanism-Informed Binding Site Intelligence
We integrate interaction fingerprints, binding site similarity, and molecular dynamics to design stable, complementary molecules with improved binding efficiency and lower attrition risk.
Deep Learning–Based Docking & Scoring
Our graph neural networks and deep learning-based scoring functions enhance pose prediction accuracy and ranking precision. Trained on extensive bioactivity datasets, these models enable nuanced understanding of molecular interactions at atomic resolution.

ADMET & Safety-Aware Optimization
Early integration of ADMET predictions, matched molecular pair analysis, and AI-based toxicity models ensures that only the most promising compounds advance to synthesis. We design for drug-likeness, metabolic stability, and safety, right from the start.
Frequently asked questions
We’re here to help with any questions you have about our plans, supported features, and how our model works.
What AI & Computational Chemistry services does PI Health Sciences offer?
PI Health Sciences provides end-to-end AI & Computational Chemistry services covering structure- and ligand-based design, virtual and ultra-large library screening, AI-driven hit-to-lead and lead optimization, active learning, and early ADMET and safety risk assessment. These services are delivered as an integrated part of active discovery programs.
How is AI & Computational Chemistry applied within real drug discovery projects?
Our AI & Computational Chemistry services are embedded directly into live discovery workflows. Computational insights are continuously reviewed by medicinal chemists and biologists, aligned with synthesis feasibility, and refined through experimental feedback, ensuring designs progress efficiently from in silico prediction to laboratory validation.
What makes PI Health Sciences’ AI & Computational Chemistry services different from standalone modeling vendors?
Unlike standalone modeling providers, PI Health Sciences delivers AI & Computational Chemistry as a chemistry-led, expert-governed service. Models operate within closed-loop discovery systems where predictions are interpretable, experimentally validated, and aligned with program objectives, development constraints, and downstream readiness.
How do these services support hit-to-lead and lead optimization programs?
Our AI-driven hit-to-lead and lead optimization services integrate generative molecular design, deep learning–based scoring, explainable AI, and structure-based modeling. This enables rapid prioritization of synthetically tractable compounds optimized for potency, selectivity, and developability.
Can PI Health Sciences support ultra-large libraries and virtual screening programs?
Yes. We provide active learning–enabled ultra-large library screening services capable of efficiently exploring million- to billion-scale chemical libraries. Adaptive prioritization strategies reduce computational and experimental burden while maintaining chemical diversity and hit quality.
Contact Us
Connect with PI Health Sciences to explore how our AI and computational chemistry capabilities can support your discovery programs, from target-driven design and virtual screening to lead optimization and safety-aware decision-making.
