AUTAC

Autophagy-Targeting Chimeras for Intracellular Protein and Organelle Degradation
At PI Health Sciences, our AUTAC Platform represents a next-generation new modality for targeted intracellular degradation, leveraging Autophagy-Targeting Chimeras (AUTAC) to address disease-relevant targets beyond the reach of conventional small molecules and proteasome-based degraders.

AUTAC technology harnesses the autophagy–lysosome pathway to selectively degrade intracellular proteins, aggregates, and damaged organelles. Unlike UPS-dependent approaches, AUTACs enable clearance of large, aggregation-prone, or proteasome-resistant substrates, expanding the therapeutic landscape for neurodegeneration, mitochondrial dysfunction, oncology, and proteinopathies.
Our integrated AUTAC discovery and development model spans hit identification through preclinical candidate nomination, combining mechanistic biology, AUTAC design chemistry, and translational pharmacology under a single scientific framework. This ensures that target selection, degradation efficiency, safety, and regulatory readiness are evaluated together, not in isolation. By integrating deep autophagy biology, rational chimera design, and IND-aligned development strategies, PI Health Sciences enables partners to advance new modality degradation programs with confidence, speed, and scientific rigor.
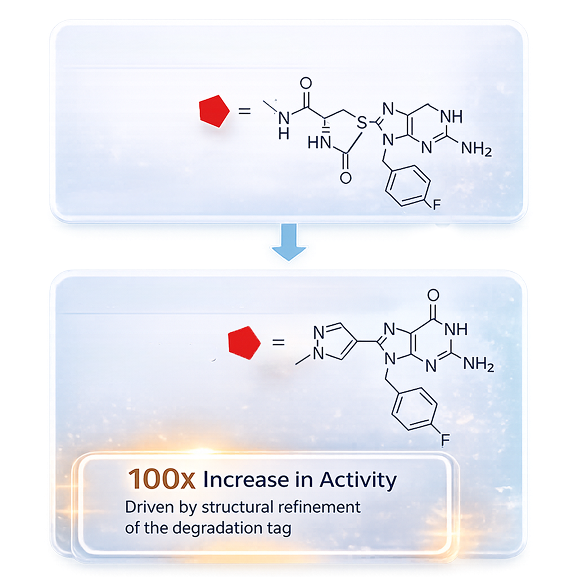
Hit-to-Candidate Workflow
Autophagy Receptors for AUTAC Development
Validated Targets & Therapeutic Applications
Our Capabilities
Expanded AUTAC Target Space
We enable degradation of aggregation-prone proteins, UPS-resistant substrates, membrane-associated targets, and damaged organelles using proteasome-independent autophagy pathways, unlocking disease biology inaccessible to conventional modalities.
Rational AUTAC Design and Chemistry Excellence
Our AUTAC design integrates warhead selection, guanine tag synthesis, and linker optimization through structure-guided chemistry to drive selective cargo recognition, autophagy engagement, and robust degradation performance.
Mechanism-Validated Degradation Framework
We deliver deep mechanistic validation through assessment of S-guanylation, K63-linked polyubiquitination, autophagy flux, receptor engagement, and cargo sequestration, ensuring pathway-specific and reproducible degradation.
Iterative Optimization and SAR-Driven Decision Making
Degradation potency, cellular efficacy, autophagy compatibility, and exposure are optimized through integrated SAR cycles supported by early PK assessment to accelerate confident lead progression.
Preclinical Translation and IND-Aligned Readiness
Programs advance through in vivo efficacy, safety and toxicology strategy, PK/PD profiling, and CMC planning to support preclinical candidate selection and seamless IND-enabling progression.
Multi-Modal New Modality Integration
AUTAC programs are strategically aligned with PROTAC, molecular glue, LYTAC, ATTEC, and AUTOTAC platforms, enabling comparative evaluation, combination strategies, and portfolio-level new modality development.
Frequently asked questions
We’re here to help with any questions you have about plans, pricing, and supported features.
How do AUTACs differ from proteasome-based degraders such as PROTACs?
AUTACs leverage the autophagy–lysosome pathway rather than the ubiquitin–proteasome system, enabling degradation of large protein aggregates, damaged organelles, and UPS-resistant substrates that are not accessible to proteasome-dependent approaches.
How is AUTAC design optimized for selective autophagy engagement?
AUTAC design integrates rational warhead selection, guanine tag chemistry, and linker optimization to induce K63-linked polyubiquitination and selective autophagy receptor recruitment, ensuring pathway-specific degradation.
What disease areas are best suited for AUTAC-based new modalities?
AUTACs are particularly well suited for neurodegenerative diseases, mitochondrial disorders, oncology, proteinopathies, and infectious diseases where aggregate clearance or organelle quality control is central to disease biology.
How does PI Health Sciences support AUTAC programs through IND readiness?
Programs are advanced through integrated optimization, in vivo efficacy, safety and toxicology planning, PK/PD profiling, and CMC strategy development, generating coherent, regulator-aligned datasets suitable for IND submission.
Contact Us
Connect with PI Health Sciences to explore how our AUTAC platform supports targeted intracellular protein and organelle degradation, from rational chimera design to IND-aligned preclinical progression.
