Molecular Glues

From Hit Identification to Preclinical Candidate
Molecular glues represent the next generation of targeted protein degradation, offering superior drug-like properties compared to bifunctional degraders. These monovalent small molecules act as chemical adhesives, inducing novel protein, protein interactions between E3 ubiquitin ligases and disease-causing proteins, leading to selective ubiquitination and proteasomal degradation.
Molecular glues are characterized by enhanced oral bioavailability, membrane permeability, and blood brain barrier penetration, along with compliance to Lipinski’s Rule of Five due to their lower molecular weight. Their simpler synthesis enables clearer structure, activity relationships and improved synthetic tractability. Clinical validation of this approach is demonstrated by FDA-approved IMiDs such as lenalidomide and pomalidomide. This modality enables access to previously undruggable targets, including transcription factors, scaffolding proteins, and neo-substrates.
At PI Health Sciences, our Molecular Glues Discovery Platform integrates hit identification technologies, AI-enabled design, and medicinal chemistry expertise to advance programs from early screening through preclinical candidate nomination. The platform supports molecular glues for targeted protein degradation with an emphasis on drug-like optimization, translational relevance, and IND-enabling readiness.
Targeted protein degradation molecule
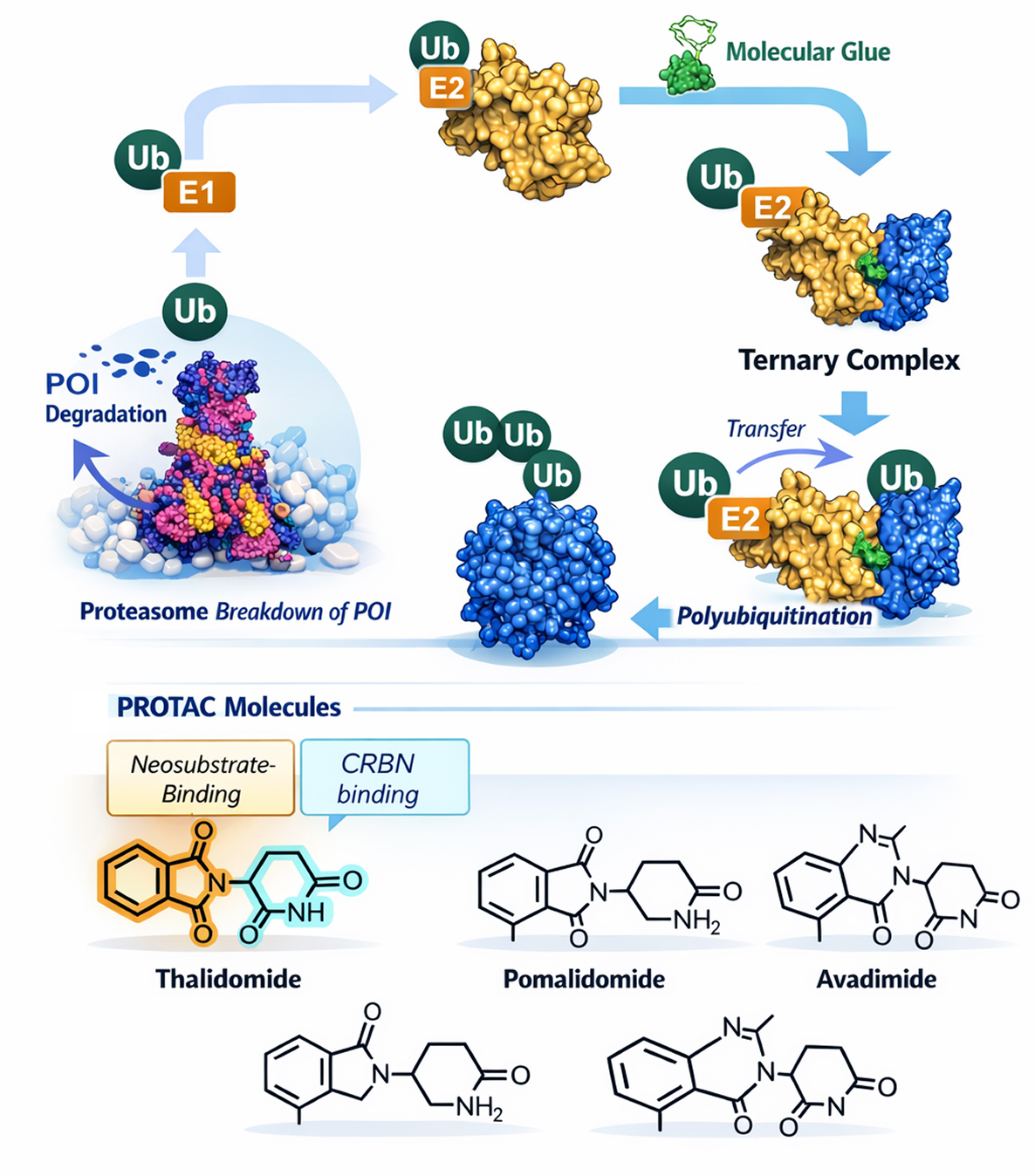
Hit-to-Candidate Workflow
E3 Ligase Focus & Validated Targets
Our Capabilities
Hit Identification
Fragment-based discovery, focused CRBN-binding libraries, IMiD-derived collections, phenotypic cellular degradation assays, binary complex screening, ternary complex screening, orthogonal confirmation.
Medicinal Chemistry Optimization
Hit-to-lead chemistry, SAR exploration, potency tuning, selectivity profiling, scaffold hopping, physicochemical property optimization, stereoselective synthesis, multi-gram scale-up.
Computational Design
Generative AI, de novo molecular design, virtual screening, molecular docking, molecular dynamics, ternary complex modeling, protein–protein interaction interface analysis.
Cellular Degradation Biology
Ternary complex formation assays, target degradation assessment, DC50 determination, Dmax determination, ubiquitination profiling, E3 ligase engagement studies, cellular target engagement assays.
ADME and Pharmacokinetics
In vitro ADME profiling, metabolic stability assessment, CYP screening, plasma protein binding, blood–brain barrier permeability evaluation, in vivo PK studies, tissue distribution analysis.
Preclinical Enablement
Exploratory toxicology, GLP toxicology planning, safety pharmacology, in vivo efficacy models, translational biomarker development, CMC development, IND-enabling package preparation.
Frequently asked questions
We’re here to help with any questions you have about plans, pricing, and supported features.
What differentiates the PI Health Sciences approach to Molecular glues (MGs)?
The platform integrates screening, computational design, medicinal chemistry, proteomics, ADME, and safety evaluation within a single workflow, enabling structured progression from hit identification to preclinical candidate selection.
How does the platform support Molecular glue optimization beyond hit discovery?
Optimization is driven by SAR, ternary complex stability, degradation kinetics, ADME and PK profiling, and in vivo efficacy data to support rational lead progression.
Which E3 ligases are supported within the platform?
The platform supports CRBN, VHL, MDM2, DCAF15, and emerging ligases, enabling access to a broad range of validated and novel neo-substrates.
How does the platform support progression toward IND-enabling studies?
Contact Us
Connect with PI Health Sciences to explore how our customized pharmaceutical development services can streamline your molecule’s journey from concept to market with precision and confidence.
